In a series of posts, we are going to talk about Mass Spectrometry.
- Introduction-The different configurations and the Electron Impact process
- What types of mass analyzers are there?
- What type of detectors are there?
- What types of analysis can be done?
- How do you read the output?
- How do they come to a qualitative measure using software?
- How do they quantitate the results?
- Do you need chromatography if you are using Mass Spectrometry?
- Other topics of interest about GC-MS
One of the limitations of any sort of detection system is the conversion of that response to a computerized system. This is called the Analog to Digital Conversion (ADC) problem. A response works in an analog domain, but computers work only in the digital domain. So, there must be a conversion. The question becomes sampling rate or the number of discrete data points that are taken. The more data points we collect the closer the digital signal is to the analog and the better the conversion is to the true signal. This whole concept is the called the data acquisition rate. This is a variable that can be controlled by software or hardware. One way we can measure is this rate is typically referred to as the number of points across (really along) the peak.
We need at least 15 to 20 points across a chromatographic peak for good quantitation. If we have fewer points we will not be able to describe the peak adequately and may lose information. For example, we may miss the top of the peak. We may miss a doublet or some other signal of co-elution. We can increase the number of points across the peak by scanning faster however data quality is sacrificed. If we increase the number of points across the peak we must increase our scan speed, so the time that we dwell or stay on and look at our mass of interest is shorter. A shorter dwell time negatively affects signal to noise (s/n). Theoretically the s/n increases with the square root of the increased dwell time.
Although the above shows the concept on LC-MS-MS, the principle holds true for GC-MS as well.
In any form of MS work, we have to remember we are “weighing” mass, not its weight. These two concepts are not the same. Some concepts are helpful as we are about to look at data. We want to define some terms so that we know how the various axes.
- The universal unit for mass is the Dalton (Da). A Dalton is equal to 1/12 the mass of the isotope of carbon 12. Stated differently 1 Da=1 atomic mass unit (amu).
- The magnitude of the charge is an electron. It is denoted as “z.” “z” is the charge of the electron generally.
- The m/z represents the daltons per unit of charge. This is the set standard that everything else is related to throughout our analysis.
The ultimate result of a hyphenated techniques such as Mass Spectrometry when it is coupled with some chromatographic technique such as Gas Chromatography or Liquid Chromatography is generally called a mass spectrum (click on to get the IUAPC). It is not a graph.
The first result will typically be a Total Ion (Current) Chromatogram. Again, it is not a graph. It is a plot along an x-axis and y-axis of the total ion current vs. retention time respectively obtained from a chromatography experiment coupled with mass detection. It is a snap shot of typically a very large window often of several hundred mass-to-charge units. (Source: Handbook of GC/MS: fundamentals and applications By Hans-Joachim Hübschmann)


- This is a TIC of an LC-MS analysis of THC and metabolites in blood: Total ion chromatogram of 10 ng/mL THC and metabolites extracted from 0.5 mL whole blood following protein precipitation, centrifugation and DPX-RP. Derivatization was performed in the CIS inlet by injecting 50 µL of DPX eluent together with 20 µL of 50/50 BSTFA/acetonitrile. No additional solvent evaporation or derivatization step was performed. Analytes: 1) THC-TMS, 2) OH-THC-TMS, 3) COOH-THC-2TMS
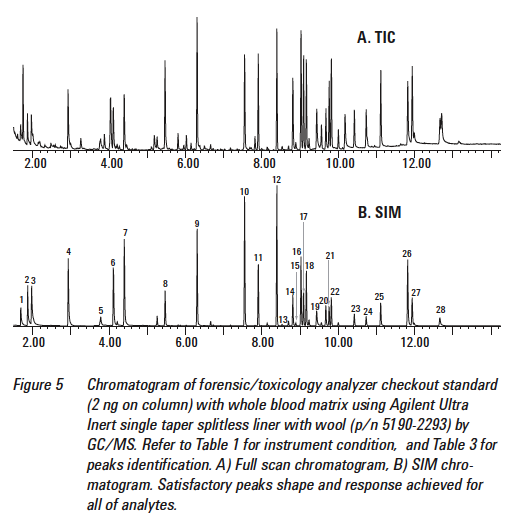
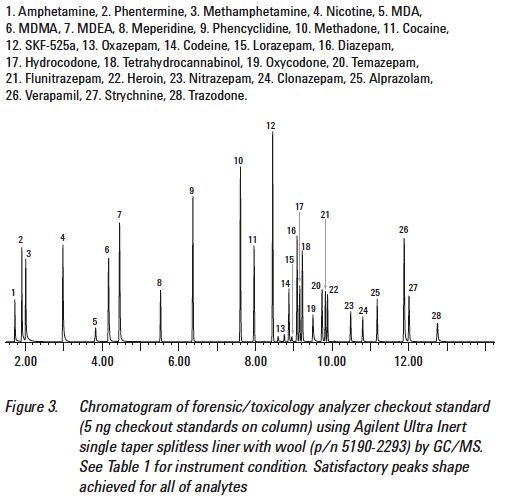
Here is a TIC from a DUID-Marijuana case.
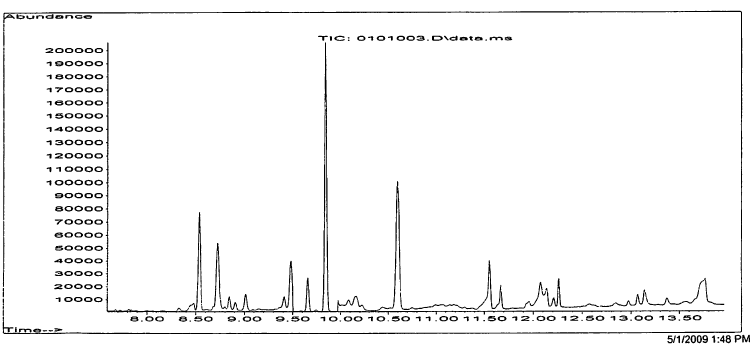
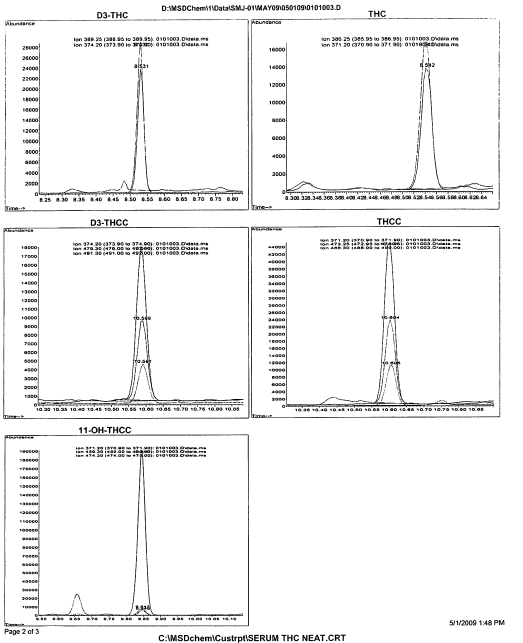
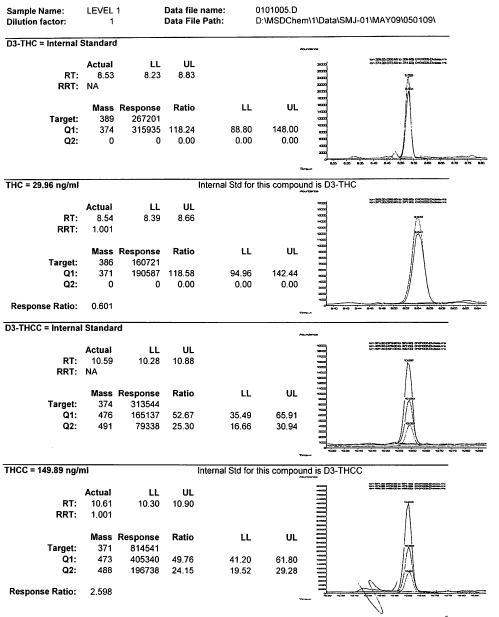
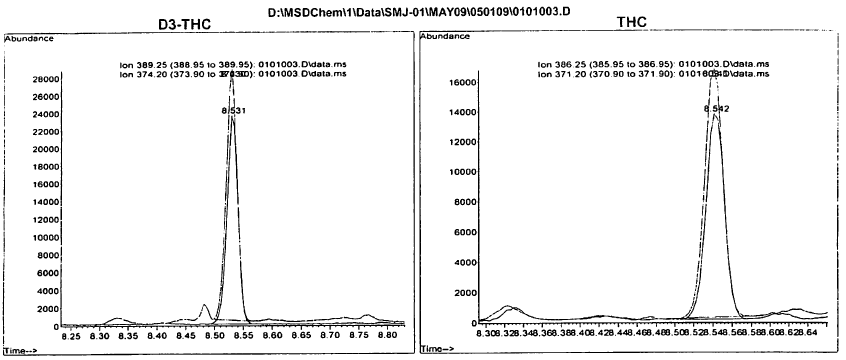
Once we have the TIC or SIM we can convert it to a mass spectrum that will then be compared against known standards (more on this later). A mass spectrum is like the below:
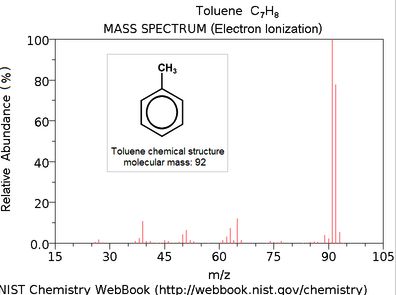
The x-axis is our old m/z. The y-axis of a mass spectrum represents signal intensity of the ions. In most forms of mass spectrometry, the intensity of ion current measured by the spectrometer does not accurately represent relative abundance, but correlates loosely with it. Therefore it is common to label the y-axis with “arbitrary units.” The highest intensity ion is assigned 100 percent with all others measured relative to it.
The very first part we should be looking for is to try to identify the molecular ion. A molecular ion is an ion formed by the removal from (positive ions) or addition to (negative ions) a molecule of one or more electrons without fragmentation of the molecular structure. The mass of this ion corresponds to the sum of the masses of the most abundant naturally occurring isotopes of the various atoms that make up the molecule (with a correction for the masses of the electron(s) lost or gained). If present then it correlates to the molecular mass of the compound.
Think of it as a boundary. In EI-based GC-MS work, if the molecular ion is present and if we have done good quality work, then we should never have a spectrum whose highest value on the x-axis exceeds the reported molecular mass.
For example, let’s use the case of ethanol. What I do is when I have data and a call by the analyst, I first take the conclusion sheet where the analyst says it is ethanol (to the conclusion of everything else in the universe), then I go to Wikipedia and some other source to find out the molecular mass. In the case of ethanol, it is reported as 46.07 g/mol or there abouts. I next go straight to the TIC, and then the scan or the SIM and determine whether or not I have isotopes reported way outside of that 46.07 range. So, for example, if I have a spectrum that produces ions at 100, then I know we need to do much further review and I am well on the track to prove the analyst’s call as wrong. Another very simple example is in the case of a spectrum that indicates a molecular weight of 18. This molecule must only contain fragments no greater than that of oxygen which has a weight of 16, hydrogen, which has a weight of 1, carbon which has a weight of 12, and/or nitrogen which has a weight of 14. We obviously turn to H2O which is 2(1)+16=18. This should be confirmed by ions at masses 17 and 16 which represent the fragmentation and logical neutral loss of HO and O.
There is a caveat to this. As GCs will only accept volatile organic compounds (VOCs), if the sample and in particular the analyte of interest is not a VOC, then we need to chemical change the sample (called derivatization) to make it capable of being analyzed by GC. What we have to remember is that not all analysis on an EI-based MS will result in a molecular ion.
In summary, when we do our analysis and we do get the molecular ion it is an important part of elucidating the unknown as it establishes the molecular weight and then often the elemental composition of the molecule (i.e., the nitrogen rule). The EI fragmentation and the corresponding EI fragment ions indicate the pieces of which the molecular ion is composed. This fragmentation is through a process called logical neutral loss analysis. It is simply a way of putting all of the pieces back to together again.



